Once the molecule file is fully loaded the image at right will become live. At that time the "activate 3-D" icon
![]() will disappear.
will disappear.
Adding
Labels and Text Annotation
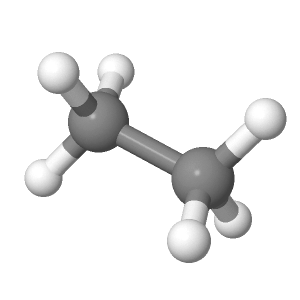
The display at right is from the file ethane-staggered-3-21G.log.
To follow along you can use the applet at right or download
the
file and open
it in a copy of the Jmol application. If you are using the applet at right you can return to this view by clicking the button below.
Getting
Started
It
is easiest to do this from the command line. Annotation using
the
menus will not be discussed in this tutorial. In both the
application
and the applet open the
script console using the pop-up menu (right
mouse button, control-click on one button mouse, > Console).
In the
application you can also use the pull-down menu (File > Script
...).
Tutorial on and more links about the Script/Console and commands.
Tutorial on and more links about the Script/Console and commands.
Topics
| >Labeling Atoms | >Labeling Groups | >Positioning Labels |
| >Basic
Floating Text (Echos) >Changing Size, Font and Color of Labels and Echos |
||
Labeling
Atoms
Select the atoms you want
to label.
- "select none" will deselect all the atoms.
- "select visible" will
select all the visible atoms.
- "select all" will select all the atoms in all frames.
- "select atomtype", where "atomtype" = carbon, hydrogen, nitrogen, etc... will select all the atoms of that type in a file.
- "select
atomexpression",
where
"atomexpression" is an expression which can be used to select
atoms within specific frames, amino acid residues and so on. Simple
examples with amino acids are shown below.
More
detailed information may be found in Bob Hanson's Jmol Scripting Documentation
 .
.
- "label" will label all selected atoms with the default information. The number following the "/" is the frame number.
- "label %a" will show just the atom symbol and index number.
- "label off" will turn off the label on the selected atoms.
- "label %e" will show just the element symbol.
- "label %P" will show the
partial charge on each element if available.
- "label "your text"" will label the atom with "your text".
- Additional options may be found in Bob Hanson's
Jmol Scripting Documentation
 under the label command.
under the label command.
The key here
is that labels are associated with particular
atoms so you must select a specific atom within the group to label.
As the example here we will use a protein/DNA complex 1lcd
(download the file to use in the application here).
To load it into the applet at right click the button below.
-
Select a particular type of atom within a group. We will select the alpha-carbon of the arginine group. The command is "select [arg]*.CA". The "*" indicates all arginines. Use the standard 3 letter abbreviations for other amino acids. The ".CA" indicates the alpha carbon. Replacing it with ".N" would select the amino group participating in the peptide bond; ".C" would select the carboxylic acid carbon participating in the peptide bond. To select all the nitrogens in all the arginines use "select (arg and nitrogen)". More detailed information may be found in Bob Hanson's Jmol Scripting Documentation
 .
. -
To label the group as arginine once you have selected only one atom in the group use "label "ARG"".
-
This label is hard to see. We can make it better by insisting it be in front of all atom (command "set labelfront") and making it a brighter color (command "color label yellow").
-
It is still hard to see which groups it is associated with. We can expand them a little by making them partially spacefilling (command: "select arg; spacefill 50%").
To start label all the hydrogens (command: "select hydrogen; label %a;")
- Center labels on atoms: "set labelalignment center; set labeloffset 0 0;" The offset is the number of pixels in the X and Y directions from the center of the atom to the beginning of the label, so the label will be aligned with the center of the atom.
- Make the labels float in front of all atoms: "set labelfront". To turn this off: "set labelfront off".
- Labels off to the left: "set labelalignment right; set labeloffset -20 0;".
- Labels off to the right: "set labelalignment left; set labeloffset 20 0;".
- To echo text in one of the nine standard positions: "set echo position; echo "your text;"", where position is: top left, top center, top right, middle left, middle center, middle right, bottom left, bottom center or bottom right. Examples: "set echo top left; echo "top left";" will display "top left" in the top left corner. "set echo bottom center; echo "bottom center";" will display "bottom center" at the center near the bottom.
- You can also use named echos so that you can move them around and do further manipulation. To name an echo issue two commands in a row:
- "set echo echoname XX% YY%", where echoname is the name you want to use for the echo and XX% and YY% are X and Y coordinates of the echo position from the bottom left in % of the applet size.
- "echo "your echo text"".
- A complete example to try is: "set echo example 50% 75%; echo "This test should appear above the ethane";"
- You can move it to a new spot with a command like "set echo example 5% 25%"
- "color label blue" colors the last labels of the currently selected atoms blue.
- "color echo blue" colors the last echo that was accessed blue.
- "font echo 24 serif bold" will make the last echo size 24 (range is 6-63), use the serif font (serif, sanserif and monospaced are the options) and make it bold faced (plain, italic and bold are the options).
- "font label 12 monospaced italic" will make the labels of the currently selected atoms size 12 monospaced italics.
Page skeleton and JavaScript generated by export to web function using Jmol 14.1.8_dev_2014.02.01b 2014-02-01 15:03: on Feb 1, 2014.
This will be the viewer

If your browser/OS combination is Java capable you will get snappier performance if you
use Java.